About the project
One of the biggest challenges in molecular biology is presented by the selection of sites for RNA splicing.
We cannot overstate its importance or apparent complexity. Splicing determines which mRNA and protein sequences a gene expresses.
It is accurate, removing introns of 102-106 bases, but paradoxically flexible in mammals, where most genes produce multiple isoforms of mRNA. These may produce proteins with different functions (up to 1,800 functional isoforms of neurexin 3, for example) and switching is involved in memory, development, differentiation, signalling and disease.
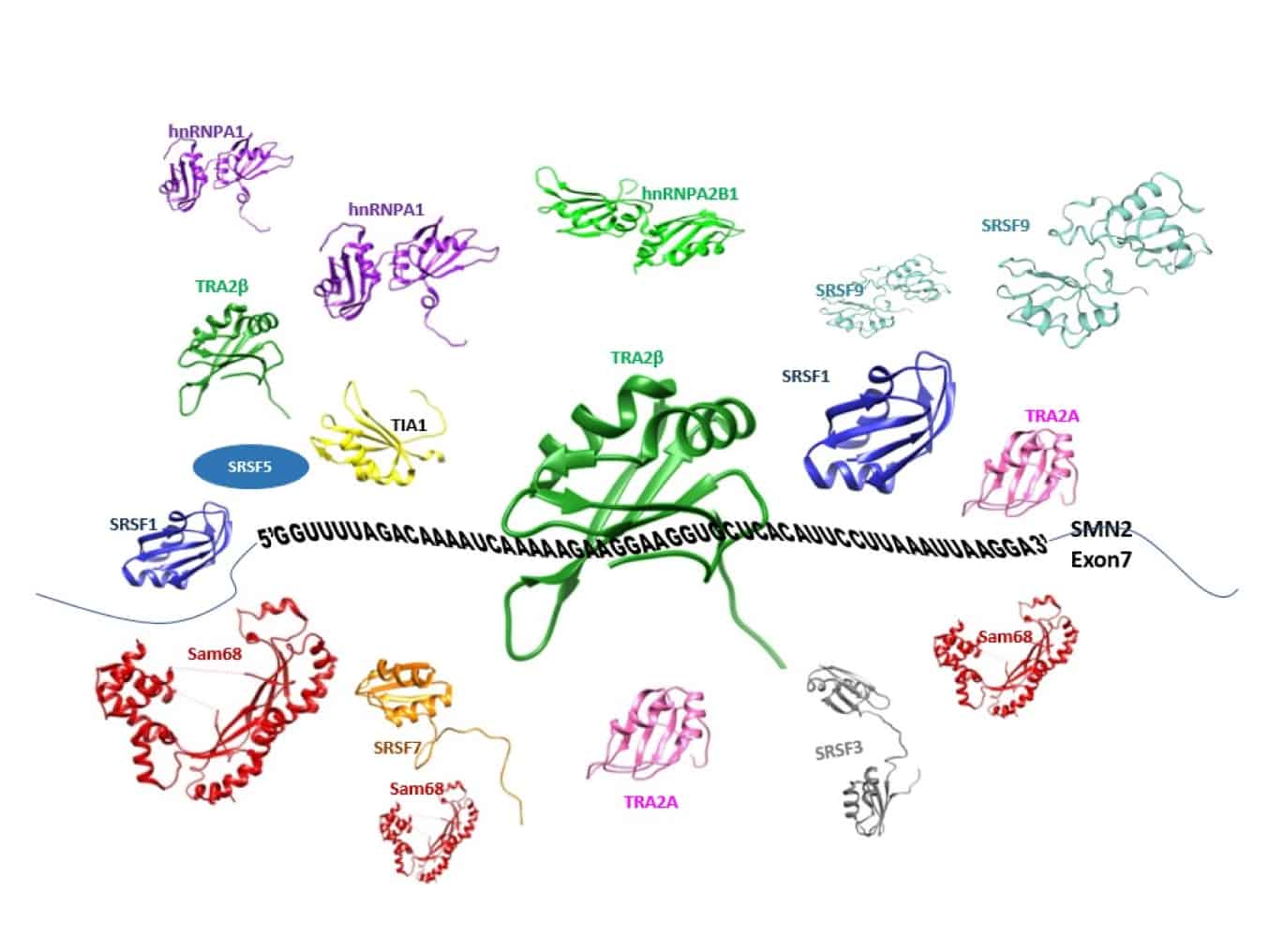
Numerous regulatory proteins and their binding sites have been identified by ensemble and ‘omic’ approaches, but our understanding of their mechanisms and functional integration remains very poor.
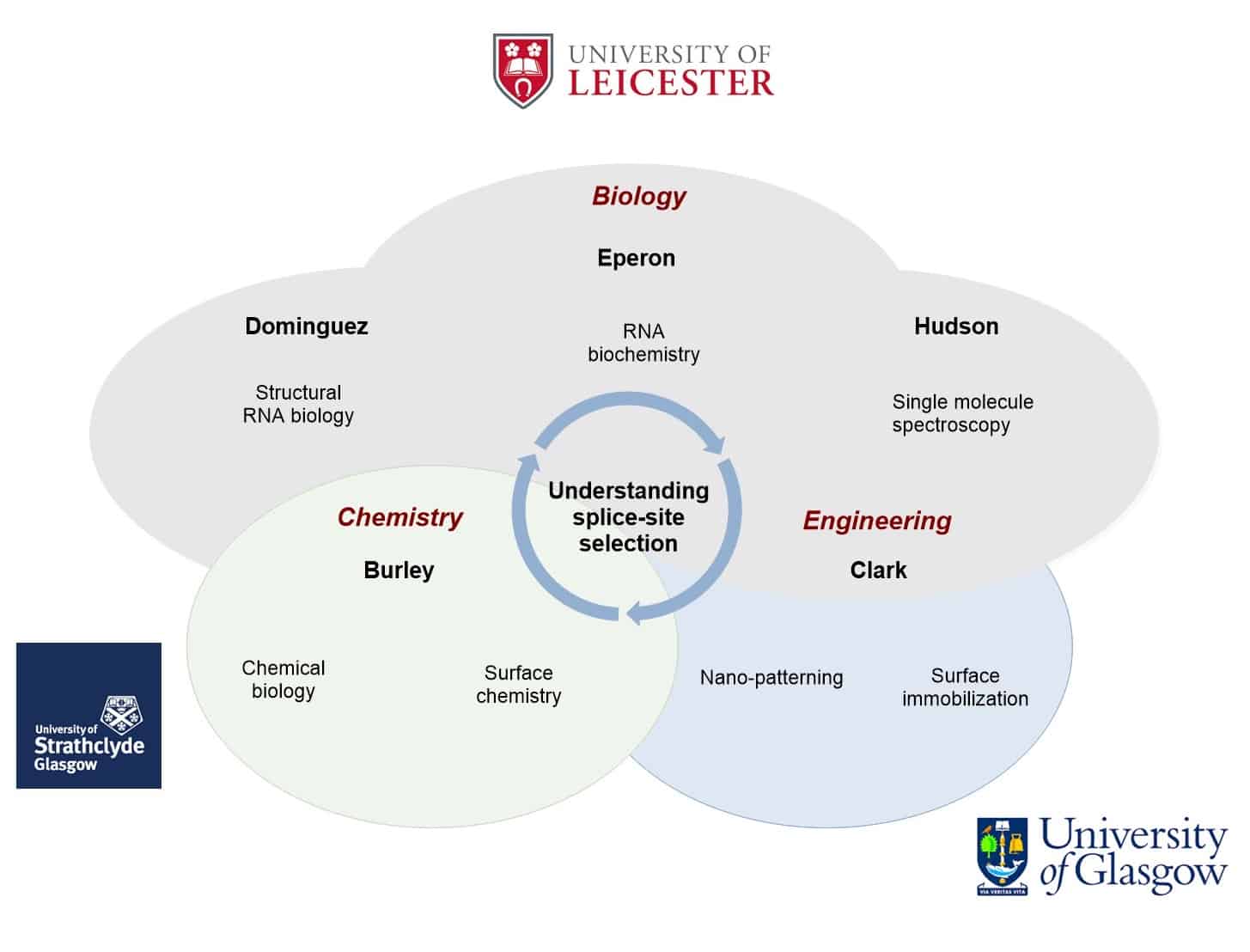
We have recently made breakthrough findings by combining single molecule methods and chemical biology.
These methods and a transformative vision of the dynamics of the process leave us uniquely able and poised to discover the mechanisms of selection.
Our multi-disciplinary work will transform our understanding of a fundamental step in gene expression, it will involve the development of new methods that should be of widespread use for many studies in the assembly of large biological complexes, and it will produce new insights that will enable researchers to develop new classes of therapeutics that re-direct and correct splicing in disease.
The purpose of this project is to find out how swarms of RNA-binding proteins, binding to many different sites on the RNA, manage to control the selection of splice sites in pre-mRNA. Selecting the right sites is essential for producing an mRNA that can be translated into protein. Moreover, the process is very flexible in humans, and switching selection to different sites allows different proteins to be expressed from a single gene. This is vital in processes ranging from memory and cell differentiation to death and disease.
This outstanding problem in gene expression can only be addressed using multi-disciplinary approaches at the molecular level. We are in a position to do this. We have developed the single molecule and chemical biology methods needed, and as a multi-disciplinary team we will complement these with structural biology and innovative new technology.
Our research will reveal the arrangements, order, dynamics, structures and interactions of these proteins. This research will be the key to understanding an entire level of gene expression, but it will also be important for the legacy of new technology and because, from an understanding of these processes, researchers will be able to develop new classes of therapeutics that re-direct and correct splicing in disease.